Importing data¶
This notebook will show you how to import a DataFrame of node attributes into Cytoscape as Node Table columns. The same approach works for edge and network attriubutes.
[5]:
import py4cytoscape as p4c
p4c.set_summary_logger(False)
p4c.cytoscape_ping()
You are connected to Cytoscape!
Always Start with a Network¶
When importing data, you are actually performing a merge function of sorts, appending columns to nodes (or edges) that are present in the referenced network. Data that do not match elements in the network are effectively discarded upon import.
So, in order to demonstrate data import, we first need to have a network. This command will import network files in any of the supported formats (e.g., SIF, GML, XGMML, etc).
[6]:
!wget https://raw.githubusercontent.com/cytoscape/RCy3/master/inst/extdata/galFiltered.sif
--2020-07-02 18:17:45-- https://raw.githubusercontent.com/cytoscape/RCy3/master/inst/extdata/galFiltered.sif
Resolving raw.githubusercontent.com (raw.githubusercontent.com)... 151.101.0.133, 151.101.64.133, 151.101.128.133, ...
Connecting to raw.githubusercontent.com (raw.githubusercontent.com)|151.101.0.133|:443... connected.
HTTP request sent, awaiting response... 200 OK
Length: 6822 (6.7K) [text/plain]
Saving to: ‘galFiltered.sif’
galFiltered.sif 100%[===================>] 6.66K --.-KB/s in 0s
2020-07-02 18:17:45 (70.3 MB/s) - ‘galFiltered.sif’ saved [6822/6822]
[7]:
!head galFiltered.sif
YKR026C pp YGL122C
YGR218W pp YGL097W
YGL097W pp YOR204W
YLR249W pp YPR080W
YLR249W pp YBR118W
YLR293C pp YGL097W
YMR146C pp YDR429C
YDR429C pp YFL017C
YPR080W pp YAL003W
YBR118W pp YAL003W
[8]:
p4c.import_network_from_file("./galFiltered.sif")
[8]:
{'networks': [51], 'views': [750]}
[9]:
p4c.export_image(filename="galFiltered.png")
from IPython.display import Image
Image('galFiltered.png')
[9]:

You should now see a network with just over 300 nodes. If you look at the Node Table, you’ll see that there are no attributes other than node names. Let’s fix that…
Import Data¶
You can import data into Cytoscape from any DataFrame in Python as long as it contains row.names (or an arbitrary column) that match a Node Table column in Cytoscape. In this example, we are starting with a network with yeast identifiers in the “name” column. We also have a CSV file with gene expression data values keyed by yeast identifiers here:
[10]:
import pandas as pd
url="https://raw.githubusercontent.com/cytoscape/RCy3/master/inst/extdata/galExpData.csv"
df=pd.read_csv(url)
[11]:
df.head()
[11]:
| name | COMMON | gal1RGexp | gal1RGsig | gal4RGexp | gal4RGsig | gal80Rexp | gal80Rsig | |
|---|---|---|---|---|---|---|---|---|
| 0 | YDL194W | SNF3 | 0.139 | 1.804300e-02 | 0.333 | 3.396100e-02 | 0.449 | 0.011348 |
| 1 | YDR277C | MTH1 | 0.243 | 2.190000e-05 | 0.192 | 2.804400e-02 | 0.448 | 0.000573 |
| 2 | YBR043C | YBR043C | 0.454 | 5.370000e-08 | 0.023 | 9.417800e-01 | 0.000 | 0.999999 |
| 3 | YPR145W | ASN1 | -0.195 | 3.170000e-05 | -0.614 | 1.150000e-07 | -0.232 | 0.001187 |
| 4 | YER054C | GIP2 | 0.057 | 1.695800e-01 | 0.206 | 6.200000e-04 | 0.247 | 0.004360 |
Note: there may be times where your network and data identifers are of different types. This calls for identifier mapping. py4cytoscape provides a function to perform ID mapping in Cytoscape:
[12]:
help(p4c.map_table_column)
Help on function map_table_column in module py4cytoscape.tables:
map_table_column(column, species, map_from, map_to, force_single=True, table='node', namespace='default', network=None, base_url='http://localhost:1234/v1')
Map Table Column.
Perform identifier mapping using an existing column of supported identifiers to populate a new column with
identifiers mapped to the originals.
Supported species: Human, Mouse, Rat, Frog, Zebrafish, Fruit fly, Mosquito, Worm, Arabidopsis thaliana, Yeast,
E. coli, Tuberculosis. Supported identifier types (depending on species): Ensembl, Entrez Gene, Uniprot-TrEMBL,
miRBase, UniGene, HGNC (symbols), MGI, RGD, SGD, ZFIN, FlyBase, WormBase, TAIR.
Args:
column (str): Name of column containing identifiers of type specified by ``map.from``
species (str): Common name for species associated with identifiers, e.g., Human. See details.
map_from (str): Type of identifier found in specified ``column``. See details.
map.to (str): Type of identifier to populate in new column. See details.
force.single (bool): Whether to return only first result in cases of one-to-many mappings; otherwise
the new column will hold lists of identifiers. Default is TRUE.
table (str): name of Cytoscape table to load data into, e.g., node, edge or network; default is "node"
namespace (str): Namespace of table. Default is "default".
network (SUID or str or None): Name or SUID of a network. Default is the
"current" network active in Cytoscape.
base_url (str): Ignore unless you need to specify a custom domain,
port or version to connect to the CyREST API. Default is http://localhost:1234
and the latest version of the CyREST API supported by this version of py4cytoscape.
Returns:
dataframe: contains map_from and map_to columns.
Warnings:
If map_to is not unique, it will be suffixed with an incrementing number in parentheses, e.g.,
if mapIdentifiers is repeated on the same network. However, the original map_to column will be returned regardless.
Raises:
HTTPError: if table or namespace or table doesn't exist in network
CyError: if network name or SUID doesn't exist, or if mapping parameter is invalid
requests.exceptions.RequestException: if can't connect to Cytoscape or Cytoscape returns an error
Examples:
>>> map_table_column('name','Yeast','Ensembl','SGD')
name SGD
17920 YER145C S000000947
17921 YMR058W S000004662
17922 YJL190C S000003726
...
Check out the Identifier mapping tutorial for detailed examples.
Now we have a DataFrame that includes our identifiers in a column called “name”, plus a bunch of data columns. Knowing our key columns, we can now perform the import:
[ ]:
p4c.load_table_data(df, data_key_column="name")
'Success: Data loaded in defaultnode table'
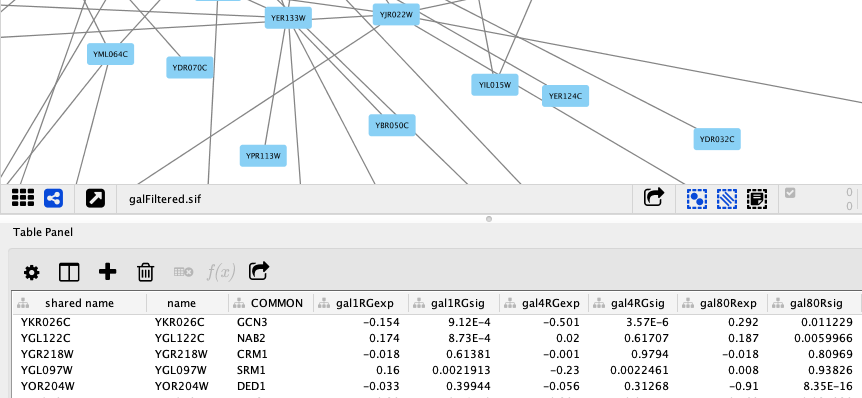
If you look back at the Node Table, you’ll now see that the corresponding rows of our DataFrame have been imported as new columns.
Note: we relied on the default values for table (“node”) and ``table_key_column`` (“name”), but these can be specified as well. See help docs for parameter details.
[ ]:
help(p4c.load_table_data)
Help on function load_table_data in module py4cytoscape.tables:
load_table_data(data, data_key_column='row.names', table='node', table_key_column='name', namespace='default', network=None, base_url='http://localhost:1234/v1')
Loads data into Cytoscape tables keyed by row.
This function loads data into Cytoscape node/edge/network
tables provided a common key, e.g., name. Data.frame column names will be
used to set Cytoscape table column names.
Numeric values will be stored as Doubles in Cytoscape tables.
Integer values will be stored as Integers. Character or mixed values will be
stored as Strings. Logical values will be stored as Boolean. Lists are
stored as Lists by CyREST v3.9+. Existing columns with the same names will
keep original type but values will be overwritten.
Args:
data (dataframe): each row is a node and columns contain node attributes
data_key_column (str): name of data.frame column to use as key; ' default is "row.names"
table (str): name of Cytoscape table to load data into, e.g., node, edge or network; default is "node"
namespace (str): Namespace of table. Default is "default".
network (SUID or str or None): Name or SUID of a network. Default is the
"current" network active in Cytoscape.
base_url (str): Ignore unless you need to specify a custom domain,
port or version to connect to the CyREST API. Default is http://localhost:1234
and the latest version of the CyREST API supported by this version of py4cytoscape.
Returns:
str: 'Success: Data loaded in <table name> table' or 'Failed to load data: <reason>'
Raises:
HTTPError: if table or namespace or table doesn't exist in network
CyError: if network name or SUID doesn't exist
requests.exceptions.RequestException: if can't connect to Cytoscape or Cytoscape returns an error
Examples:
>>> data = df.DataFrame(data={'id':['New1','New2','New3'], 'newcol':[1,2,3]})
>>> load_table_data(data, data_key_column='id', table='node', table_key_column='name')
'Failed to load data: Provided key columns do not contain any matches'
>>> data = df.DataFrame(data={'id':['YDL194W','YDR277C','YBR043C'], 'newcol':[1,2,3]})
>>> load_table_data(data, data_key_column='id', table='node', table_key_column='name', network='galfiltered.sif')
'Success: Data loaded in defaultnode table'
[ ]: